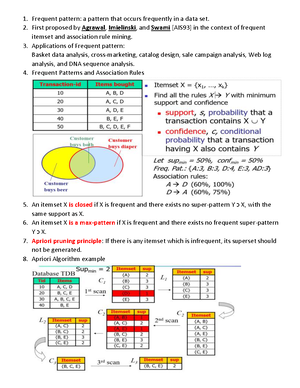
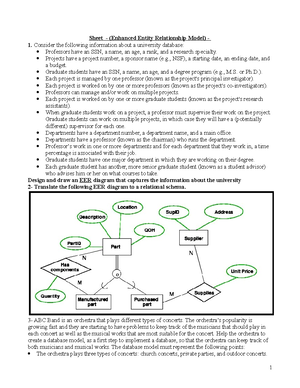
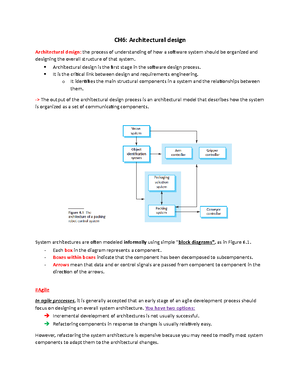
- Information
- AI Chat
Data Mining - Biological data analysis Lecture 1
Data Mining
Assiut University
Related documents
- Data Mining - Ch10 Mining texts and web data Lecture 2
- Data Mining - Ch10 Mining texts and web data Lecture 1
- Data Mining - Ch10 Mining Object, Spatial, and Multimedia Data Lecture 4
- Data Mining - Ch10 Mining Object, Spatial, and Multimedia Data Lecture 2
- Data Mining - Ch10 Mining Object, Spatial, and Multimedia Data Lecture 3
- Data Mining - Ch9 Multirelational Data Mining Lecture 3
Preview text
Data Mining Concepts
Bio Mining- Lecture 5 : Biological Data Analysis
Topics
- We will explore the syllabus
through a series of questions?
- Please ASK
- All logistical information will be
given at the end
Life begins with Cell
•A cell is a smallest structural unit of an organism that is capable of independent functioning •All cells have some common features
All life depends on 3 critical
molecules
•Protein –Form enzymes, send signals to other cells, regulate gene activity. –Form body’s major components (e. hair, skin, etc.).
- DNA –Hold information on how cell works
•RNA –Act to transfer short pieces of information to different parts of cell –Provide templates to synthesize into protein
History of Genbank
•In 1982 Goad's efforts were rewarded when the National Institutes of Health funded Goad's proposal for the creation of GenBank, a national nucleic acid sequence data bank. By the end of 1983 more than 2,000 sequences (about two million base pairs) were annotated and stored in GenBank.
Sequence data
Sequence data refers to a type of data that is ordered in a specific sequence or pattern. This type of data is commonly found in various fields such as genetics, finance, and natural language processing. Examples of sequence data include DNA sequences, stock market prices over time, and sentences in a paragraph. The analysis of sequence data often involves techniques such as pattern recognition, time series analysis, and machine learning algorithms to identify trends and patterns within the data.
How do we query a
sequence database?
•By name
•By sequence
•‘Relational’ queries are barely applicable
Quiz:DNA sequence
databases
§Suppose you have a 100nt sequence, and you want to know if it is human, what will you do? §How much time will it take? Or, how many steps? (Query=m, Database = n) •What if you were interested in identifying the human homolog of a mouse sequence ( 85% identical)? How much time will it take? What if the query was 10Kbp? What if it was the entire genome?
database ACGGATCGGCGAATCGAATCGTGG GCCTTA
query AATCGT
BLAST
•Allows querying sequence databases with sequence queries.
Quiz:BLAST
§What do you do if BLAST does not return a ‘hit’?
§What does it mean if BLAST returns a sequence that is 60% identical? Is that significant (are the sequences evolutionarily related)? §Suppose Protein sequences A & B are 40% identical, and A &C are 40% identical. If we know that A&B are evolutionarily related, what does that say about A & C?
Non sequence based
queries
•Biological databases are not
limited to sequences.
Non-sequence based queries refer to a type of query that does not require the data to be in a specific order or sequence. In other words, these queries can retrieve information from a database without relying on the order in which the data was entered. Non- sequence based queries are often used in databases that contain large amounts of unstructured data, such as text documents or multimedia files. These queries can be more flexible and efficient than traditional sequence- based queries, as they allow for more complex searches and analysis of data.
Protein Sequences have
structure
Can you search using a structure query?
Yes, I can search for Protein Sequences that have structure using a structured query. To do this, I would need to use a database or search engine that allows for structured queries, such as the Protein Data Bank (PDB). Within the PDB, I could use a query language such as SQL to search for Protein Sequences that
have structure by specifying certain criteria such as the presence of certain amino acids or structural motifs. Alternatively, I could use a tool like BLAST to search for similar sequences in the PDB and then filter the results based on whether or not they have known structures.
important to choose the appropriate tool based on your specific research question and the type of data you are working with.
- What if the database was a collection of patterns?
If the database was a collection of patterns rather than protein sequences, you could still use similar bioinformatics tools to search for matches or similarities between your input pattern and the patterns in the database. However, it may require some additional preprocessing or conversion of the data to make it compatible with these tools.
Database of Protein
Motifs
Quiz: Protein Sequence
Analysis
Proteins fold into a complex 3D shape. Can you predict the fold by looking at the sequence?
Proteins are known to fold into a complex 3D shape, which is essential
Data Mining - Biological data analysis Lecture 1
Course: Data Mining
University: Assiut University

- Discover more from:Data MiningAssiut University91 Documents
- More from:Data MiningAssiut University91 Documents
Recommended for you
Students also viewed
Related documents
- Data Mining - Ch10 Mining texts and web data Lecture 2
- Data Mining - Ch10 Mining texts and web data Lecture 1
- Data Mining - Ch10 Mining Object, Spatial, and Multimedia Data Lecture 4
- Data Mining - Ch10 Mining Object, Spatial, and Multimedia Data Lecture 2
- Data Mining - Ch10 Mining Object, Spatial, and Multimedia Data Lecture 3
- Data Mining - Ch9 Multirelational Data Mining Lecture 3